psc systems power dna-based identification of surface, airborne microbes worldwide
Samples from 60 cities reveal previously unknown microbes, diversity of antibiotic-resistance genes
by Ken Chiacchia
Dangerous microbes can emerge with little warning. Using PSC’s advanced research computers, a team of scientists led from Weill Cornell Medicine has undertaken a vast analysis of microbial DNA in thousands of urban air and surface samples worldwide. The results revealed city-specific “fingerprints” of bacteria and viruses. They also gave us a first look at the population of dangerous antibiotic-resistance-conveying genes across the globe, as well as thousands of previously undiscovered species in the urban microbial world.
Why It’s Important
The rise of COVID-19 has given the world a harsh lesson on the importance of being aware of what the microbial world is doing, and which bacteria and viruses are where. Public health experts would like to monitor the microbial world much as an airport control tower monitors the airspace around it, knowing which aircraft are near and where they’re heading.
“In a nutshell, we wanted to build a genetic, functional and geospatial map of the DNA world’s cities, just kind of like the Google Maps of DNA of the Earth.” -Christopher Mason, Weill Cornell Medicine
That’s why a huge international collaboration, led by Christopher Mason of Weill Cornell Medicine, decided to employ the immense power of advanced research computing to assemble the DNA sequences of bacteria and viruses in the air in six cities and on surfaces in public transit locales in 60 cities worldwide. Using PSC’s three most powerful supercomputers in three different eras—Blacklight from 2010 to 2015, Bridges from 2015 to February 2021, and Bridges-2 since—the team sequenced thousands of microbes’ DNA all at once, using the computers’ brute force to sort the genes and species electronically into a many-species metagenomic map for use by scientists and public health experts.
How PSC Helped
Many of the most-used DNA sequencing methods can sequence at most a few hundred nucleotides—the A, C, T, G alphabet of the genetic code—at a time. Because of that, scientists need to match overlapping fragments of DNA to put the millions of nucleotides in an organism’s genome in proper order. When the task is to sort and assemble DNA fragments from thousands of species of bacteria and viruses at once in a sample from the environment, this assembly task becomes enormous. Blacklight, Bridges and Bridges-2 all offered large-memory nodes that made this kind of task possible. The same as RAM in a personal computer, larger memory allows the machine to compare more fragments at once without wasting time going back to storage—like in a PC’s hard drive—for more data.
Graduate student David Danko at Weill Cornell Medicine and research associate Daniela Bezdan at Weill Cornell Medicine and the Abdulaziz Alsaud Institute for Computational Biomedicine worked with Mason and hundreds of scientists worldwide to collect 4,728 surface samples from mass transit locations in 60 cities worldwide in 2016 and 2017, and analyze their DNA sequences using PSC’s systems. In parallel work, M. H. Y. Leung and X. Tong at the City University of Hong Kong and K. O. Bøifot at the Norwegian Defence Research Establishment performed a similar analysis of 259 airborne samples in Denver, Hong Kong, London, New York City, Oslo and Stockholm.
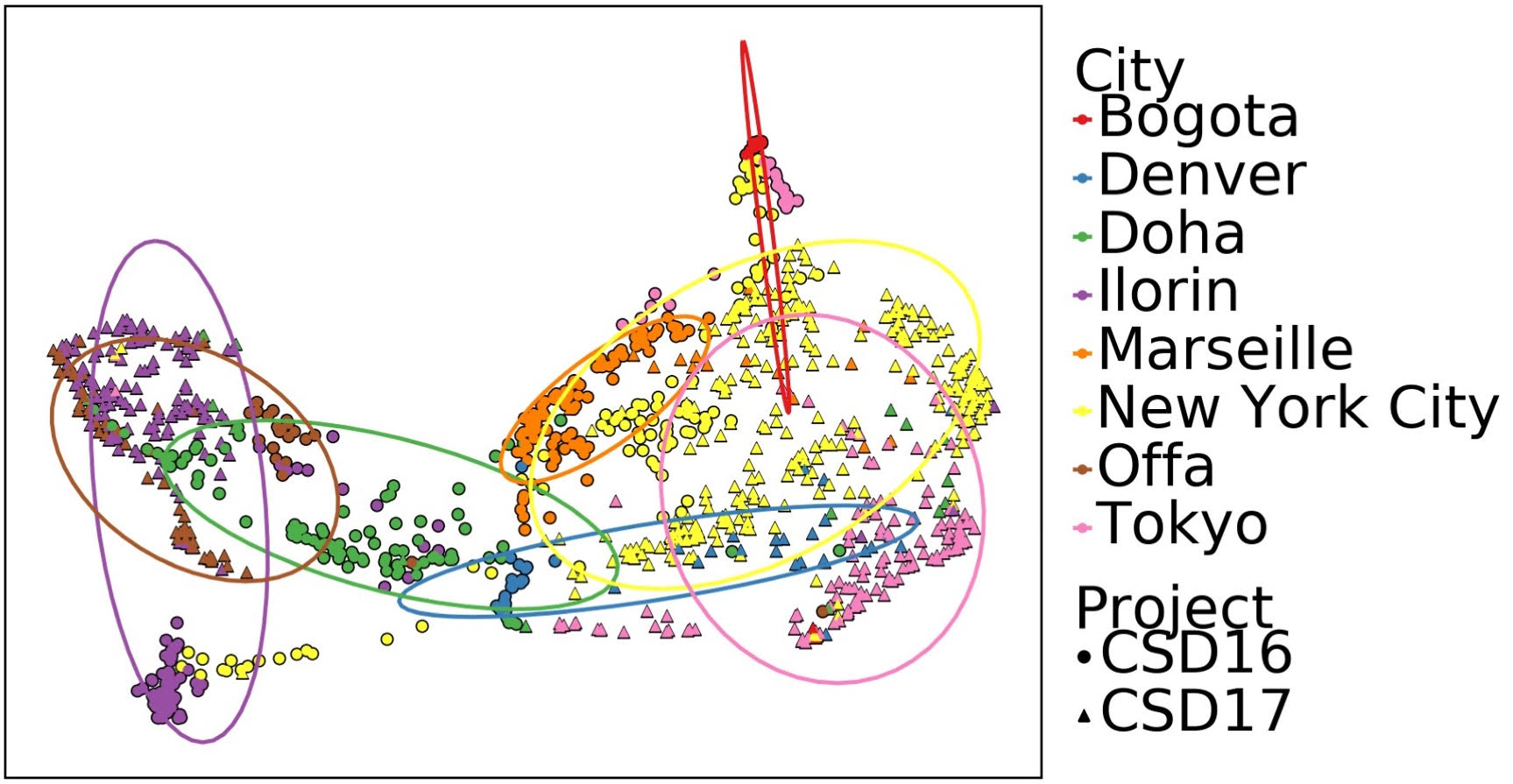
The fingerprint of the microbial species in a given city’s public transit surfaces (coded by color) changed over time (samples from 2016 shown as circles, 2017 triangles). But the cities remained distinct and different from each other. From Danko D, Bezdan D et al. “A Global Metagenomic Map of Urban Microbiomes and Antimicrobial Resistance,” Cell, May 27, 2021.
“To do all of our de novo assembly at scale, [PSC’s systems] gave us the fastest and most expansive computational framework through which we could assemble all the sequences to find what were the real novel species and the novel genetic elements in this data set … It probably literally wouldn’t have been possible in this time frame without that infrastructure … Also with these assembly projects, we will hit, occasionally, issues with bugs in the code or challenges with file structures. And Phil [Blood, PSC senior director of research] was a key and a pivotal collaborator to make sure that we can actually do all the assemblies and get them up and running well … He’s our go-to man for ‘What happened, what seems to be breaking?’”—Christopher Mason, Weill Cornell Medicine
Chris Mason, Weill Cornell Medicine
The assembly results gave scientists their first metagenomic map of urban areas worldwide, opening a new era of disease surveillance. The surface samples contained more than 15,000 species of virus, bacteria and archaea, primitive bacteria-like organisms from which more complex plants and animals evolved. The airborne samples showed evidence of more than 450 microbial species. More interesting, fewer than 10 percent of the microbes identified from their DNA were species known to science, revealing a vast unknown microbial environment.
As expected from earlier research, cities had distinct microbial populations, with varying amounts of 31 species from surfaces and 17 from the air forming a kind of fingerprint that the scientists could use to identify the city of origin. These fingerprints varied over time, though the cities remained recognizably different. The team’s results suggest that differences in climate, geography, population density and other factors may help drive these distinctions. One important facet of the work was to detect and monitor differences in 20 known genes that give bacteria resistance to antibiotics. These also differed widely between the cities.
Together, the results offer the first high-resolution view of the types of microbes that exist in the environment in a way that can be harnessed to public health efforts. The collaborators reported their results in two papers, a coveted cover story in the journal Cell on May 26, 2021, and an upcoming report in the journal Microbiome. The team is expanding their research, now collecting RNA data, which will open up a view of RNA viruses such as the coronavirus that causes COVID-19. They’re also investigating artificial-intelligence driven classification of the results, to automatically detect metagenomic shifts that pose a threat to human health.

