Computation with Bridges Points to Unexpected Mechanism for DNA Duplication in Parkinson’s Disease
The progressive disability of Parkinson’s disease carries a heavy toll both in numbers—nearly a million people in the U.S. have it—and in suffering. The wrong number of copies of a gene called SNCA is one cause of Parkinson’s. Working with an international collaboration, scientists from the University of Texas MD Anderson Cancer Center used PSC’s Bridges platform to study the DNA sequences surrounding SNCA-gene duplications in Parkinson’s patients. Their work identified an unexpected mechanism for the mutations. This discovery could lead to better diagnosis, prediction of disease progression and even treatments.
Why It’s Important
Nearly one million people in the U.S. live with the progressive disability of Parkinson’s disease. The condition costs $2,500 per patient annually for medications, with as much as another $100,000 for surgery. But those numbers can’t completely capture the human cost of progressively losing the ability to walk and stand, to talk or to write and, sometimes, ending in dementia and hallucinations.
Parkinson’s develops in a complicated interaction between environmental causes and changes in the genes. About 5 percent of Parkinson’s patients have a mutation in the SNCA gene, which produces a protein called alpha-synuclein. When in excess, this protein accumulates in tangles in brain cells, killing the cells and causing the disease symptoms. Normally people have two copies of the SNCA gene, one from each parent. But sometimes the DNA gets scrambled, deleting or duplicating those genes. While two copies function correctly, too few or too many can lead to Parkinson’s.
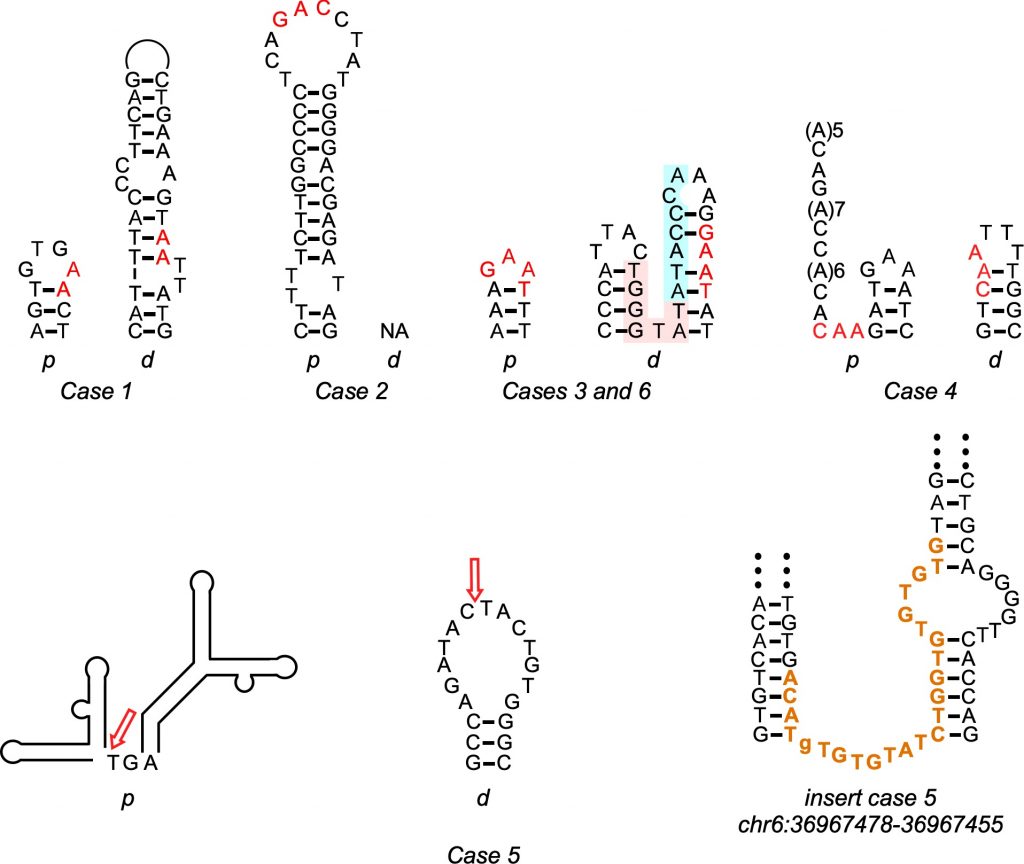
“In this particular case we have six individuals who have Parkinson’s because they have a segment of the DNA that is duplicated in chromosome 4. Although the length of the segment varies, in all cases [it involves] one gene—SNCA, which is expressed in the brain and also … has to be very tightly regulated for the cells to work well.”—Albino Bacolla, University of Texas MD Anderson Cancer Center
A better understanding of how these DNA rearrangements happen could help doctors better predict patients’ course of disease and possibly even lead to treatments. Albino Bacolla, working in the laboratory of John Tainer at the University of Texas MD Anderson Cancer Center and with an international team led by the Seoul National University Hospital in South Korea, used PSC’s National Science Foundation-funded Bridges supercomputing platform to uncover the clues to this process hidden in the genes of six Parkinson’s patients with duplications of their SCNA genes.
How PSC Helped
DNA can be a fragile molecule. Since it carries the genetic information needed to accomplish life processes, it’s critical for living things to repair it when it’s damaged. In particular, breaks in either one of both of the DNA strands must be rejoined for the cell to survive.
To repair such damage, the cell needs to know what the correct sequence in the repaired DNA strand should be. Because DNA is double-stranded, each strand has a matching sequence on its opposite strand. The cell uses the opposite strand as a guide for the repair. But sometimes the sequence of a DNA strand allows it to pair with itself in a loop instead of pairing with its opposite strand. This hides it from the repair machinery, which skips to another place in the DNA, even on another chromosome. Such mistakes can delete or duplicate genes.
Doctors had long suspected a particular kind of faulty repair called nonhomologous end joining (NHEJ) in the duplications seen in SNCA mutations. That’s when the cell attaches a break in the DNA to another place in the DNA, regardless of the sequence in the original and new strand. But a number of other mechanisms were possible. One is that the cell finds a relatively long match in sequence to the damaged DNA elsewhere and rejoins it there. That process is called nonallelic homologous recombination (NAHR).
Bacolla used Bridges’ ability to match many DNA sequences simultaneously to compare the patients’ genes to each other and to a reference database that contained “normal” DNA sequences. The Big Data task took about one week on Bridges to complete; a similar computation could have taken months on a personal computer.
“We just can’t look at these data and really understand them without the power of advanced computing. We really needed the computational power and tools of Bridges to make any sense of it.”—John Tainer, University of Texas MD Anderson Cancer Center
The Bridges computations supported something of a dark-horse candidate for the Parkinson’s mutations. The duplicated genes were flanked by regions of repeated DNA sequence, which was highly unlikely if the random rejoining of NHEJ were happening. But the repeats were short, which was unlikely to happen in NAHR. That’s because NAHR requires an extensive matching sequence, which is preserved at each end of a duplication after the repair. Instead, the short DNA repeats on either side of the duplicated genes were most consistent with microhomology-mediated break-induced replication, or MMBIR. The scientists reported their results in a paper in the journal Movement Disorders in May 2020.
Identifying MMBIR as the most likely cause for the Parkinson’s mutations poses some interesting follow-on questions. Is this method of repair more common than scientists had thought? Can it form the basis for a better understanding of such mutations that can lead to improved diagnoses of which people at risk for Parkinson’s are most vulnerable? Of course, the possibility that the mechanism of such mutations can offer clues for reversing or treating the condition in patients is also a major future topic for research.
You can read the Movement Disorders paper here.