CRISPR Simulation Points to Improved DNA Editing, Gene Therapy
Virtual Cas9 Protein on Anton 2 at PSC Aids Search for Cures for Genetic Diseases
by Ken Chiacchia
The CRISPR-Cas9 system from bacterial proteins is the basis for a promising method for editing DNA. The method offers cures for genetic diseases, but sometimes causes “off-target” DNA changes and possibly serious side effects. A team from the University of California Riverside is using the D. E. Shaw Research (DESRES) Anton 2 supercomputer hosted at PSC to show how binding to the target DNA activates the Cas9 protein’s DNA-cutting function. They hope to use the information to design more accurate CRISPR tools without such side-effects.
Why It’s Important
Since the discovery of how mutations in the DNA can cause genetic disease, doctors have wanted to be able to snip out and correct the errors. A single change in the DNA code affects people with some types of inherited high blood cholesterol, sickle cell anemia, polycystic kidney disease, cystic fibrosis, Tay-Sachs disease, Duchenne muscular dystrophy, hemophilia, Huntington’s disease and others. Editing the DNA could Cure—with a capital “C”—those diseases, not just treat them.
That’s the reason for the excitement over the discovery of the CRISPR-Cas9 system in bacteria. The natural form of the protein acts as a primitive immune system, teaming up with a guide RNA molecule to add a target DNA sequence to incoming virus genes. This target marks the virus DNA for destruction, protecting the bacterium from the virus. Scientists learned how to alter the Cas9/guide RNA to make their own changes to DNA, for the first time offering the hope of precise, side-effect free gene therapy. The technology is at the center of a number of experimental treatments for genetic disease now being tested.
“We like to say that the Cas9 protein is a little bit like an aerialist artist! Indeed, Cas9 binds the guide RNA and then grabs a matching DNA through a fascinating dance … but the changes in conformation to allow this binding are unknown. How does it work? In this sense, molecular dynamics simulations have the unique capability to capture this dance at the atomic level.”—Giulia Palermo, University of California Riverside
Cas9 is much more precise and accurate in changing the DNA than previous technologies. But it isn’t perfect. It can sometimes make off-target changes to the DNA. At best, these changes don’t help. At worst, they could cause serious side effects like cancer. That’s why Giulia Palermo and her team of scientists at the University of California Riverside decided to study how the protein works, down to the nuts and bolts. If they could understand Cas9’s details, they reasoned, they could find ways of engineering it to be even more precise. Their studies required the longest, most accurate computer simulations of Cas9 possible.
The team, consisting of Palermo, post-doctoral researchers Lukasz Nierzwicki and Pablo Arantes, and students Aakash Saha and Rohaine V. Hsu, turned to Anton 2, a special-purpose supercomputer developed and made available without cost for non-commercial research by DESRES and hosted by PSC with support from the National Institutes of Health. As a post-doctoral researcher in the lab of J. Andrew McCammon at the University of California San Diego, Palermo had pioneered computational studies of CRISPR-Cas9 using Anton 2 and a novel Gaussian accelerated Molecular Dynamics (GaMD) method using other supercomputers.
How PSC Helped
The team wanted to understand the complicated series of motions that the Cas9 protein has to undergo to attach to and cut DNA. Previous computer simulations and lab results hinted that the process would take several steps.
Cas9 consists of a number of domains, or functional parts. Two domains, called REC1 and REC2, hold onto the guide RNA and match it to the DNA that the protein means to cut. But at this point, the domain that actually does the cutting, called HNH, is stuck in an inactive state. The team had previously used GaMD on other supercomputers to try to figure out how this inactive state works, and how the protein reverses it so that HNH can cut the DNA.
The work showed that it would be possible to observe the activation of HNH, but only if the team could simulate the system for much more virtual time. The process had previously been simulated for 400 nanoseconds—billionths of a second. Anton 2, because it is designed specifically for such molecular dynamics simulations, can simulate complex molecules for much longer than general-purpose supercomputers. The DESRES supercomputer provided the more than about 16 microseconds of simulated time—40 times the previous duration—needed to work out the details.
“If Anton wouldn’t have been available, I think that this amount of sampling wouldn’t have been possible. Using earlier simulations, based on a novel Gaussian accelerated molecular dynamics method, we saw some movements that actually told us that this would have been possible to simulate. However, perhaps without Anton we wouldn’t have been able to observe the full conformational activation in a timely fashion.”—Giulia Palermo, University of California Riverside.
Using Anton 2, the team was able to observe the activation of HNH, showing how it approaches the DNA for cleavage in “real time” and at the level of individual atoms in the protein. The Anton 2 simulations confirmed the earlier predictions, showing in detail how the intricate dance of Cas9’s parts coordinate to recognize, bind and clip the DNA. The scientists reported their results in the journal Quarterly Reviews of Biophysics in 2018, with another article that surveyed their understanding of the protein at a higher level in the journal WIREs Computational Molecular Science in July 2020. You can read these papers here and here. Better, recent lab results with cryo-electron microscopy have shown that the protein moves the same way in the test tube as predicted using Anton 2, at a high level of agreement.
The UC Riverside team is now using Anton 2 as well as other computing tools to determine how the protein creates off-target effects. They’re also using the DESRES supercomputer to study the related CRISPR-Cas12 protein, another bacterial defense protein that has been recently harnessed for a new type of quick test for the SARS-CoV-2 virus that causes COVID-19. The UC Riverside team involves two talented students who are pursuing their graduate studies in computational biophysics. More information on how this group is teaming to define the dynamics and mechanism of CRISPR systems can be found here.
Animation of the molecular dynamics simulation showing the intricate dance by which the Rec1, Rec2 and Rec3 domains of Cas9 coordinate to activate the HNH domain’s DNA-clipping function.
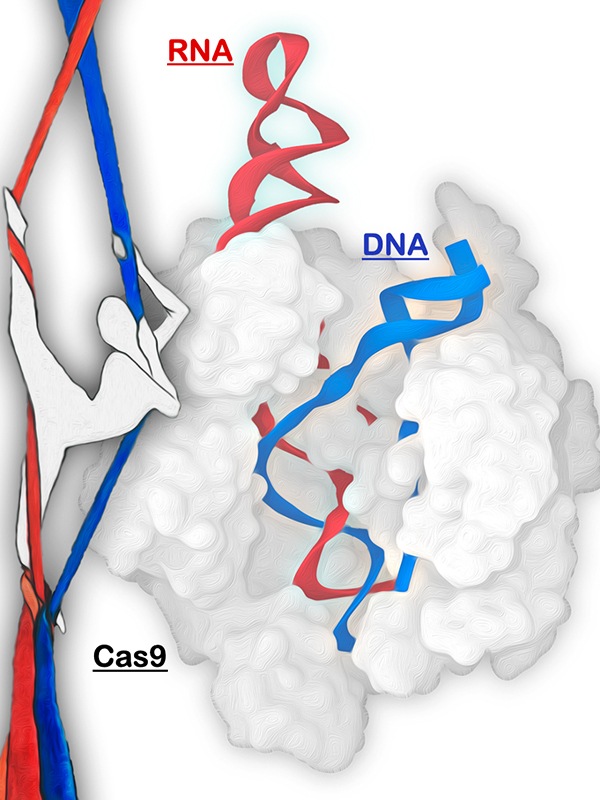
The CRISPR-Cas9 genome editing system is represented as an aerial silk artist. The Cas9 protein (gray) is shown as a ballerina that grabs the RNA (red) to recruit the DNA (blue). Original artistic rendition by Amelia Palermo. Graphical manipulation by Clarisse G. Ricci.
Giulia Palermo, University of California Riverside