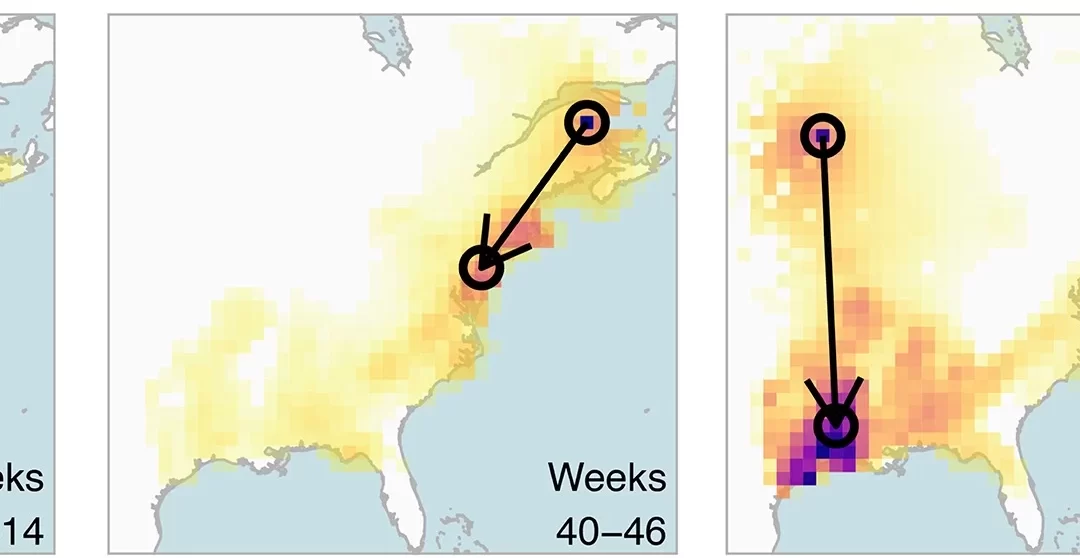
by Ken Chiacchia | May 10, 2023 | AI, Science Highlights
Predictions made by the BirdFlow AI on how the American woodcock migrates in North America between eBird “snapshots.” From Fuentes, M., Van Doren, B. M., Fink, D., & Sheldon, D. (2023). BirdFlow: Learning seasonal bird movements from eBird data. Methods in Ecology...
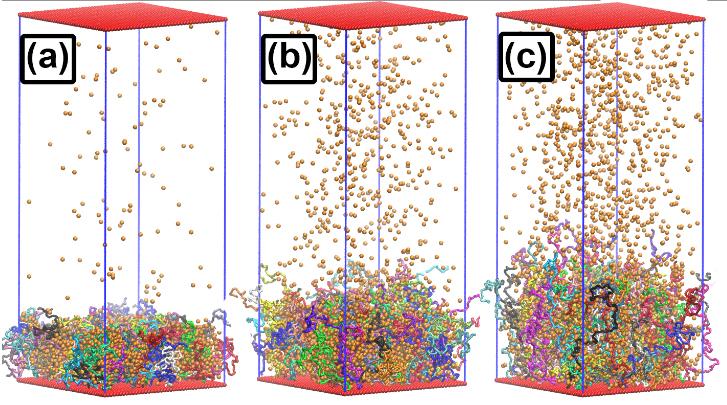
by Ken Chiacchia | Apr 26, 2023 | Bridges-2, Science Highlights
Proof-of-concept work suggests applications in industrial processes, environmental monitoring, and medicine Polymer brushes are forests of molecular-sized chains extending from a surface. If they can be made to behave properly in the real world, they could form the...

by Ken Chiacchia | Apr 12, 2023 | Bridges-2, Science Highlights
Work on Bridges-2 shows the approach can help explain persistence of some cultural practices as well as how culture changes over time Do human culture and practices evolve in a predictable way over time? A team from Carnegie Mellon University and the Santa Fe...

by Ken Chiacchia | Mar 29, 2023 | Science Highlights
Adobe Stock #527516305. Scientists leverage past MARC program to maintain competitive bioinformatics program at minority-serving institution Lucina pectinata is a clam that lives in mud flats among levels of hydrogen sulfide high enough to harm most animals....
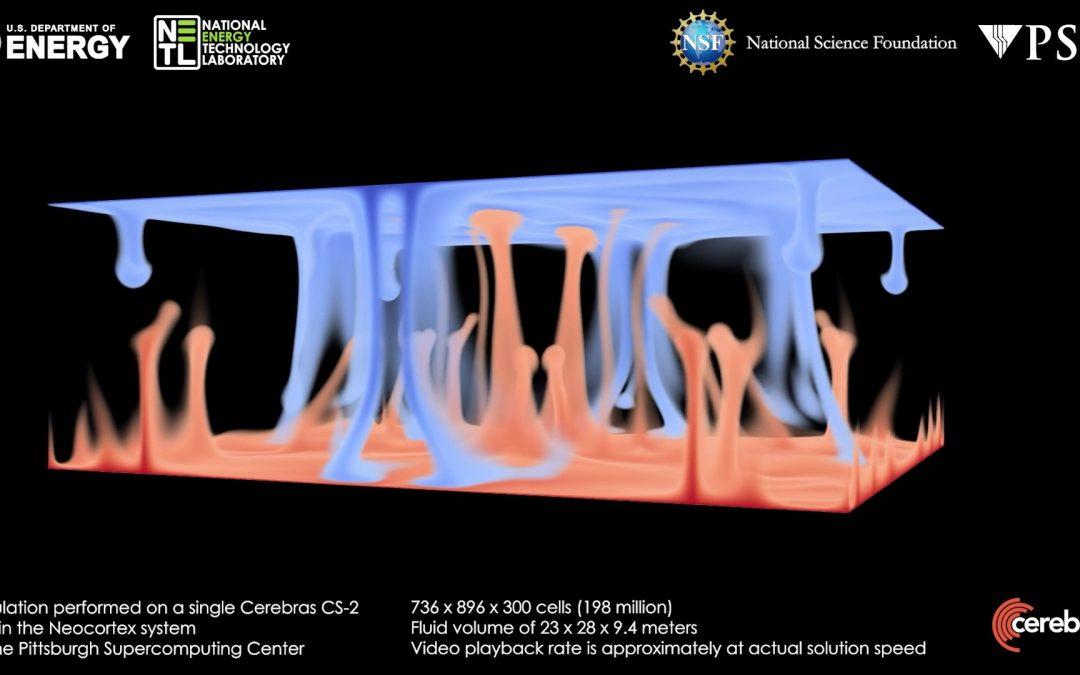
by Ken Chiacchia | Mar 15, 2023 | Neocortex, Science Highlights
Neocortex-computed still-image from a simulation of the classic Rayleigh-Bénard convection problem, which shows how a fluid layer begins mixing as it’s heated from the bottom (red) and cooled from the top (blue) in a gravitational field. Real-time simulations offer...
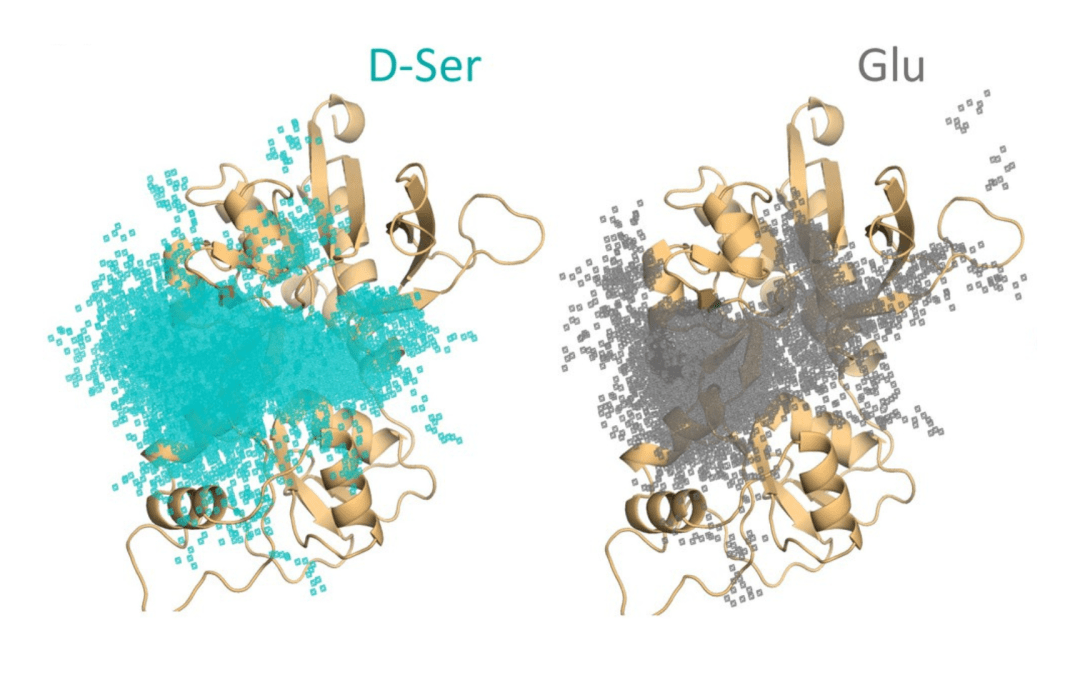
by Ken Chiacchia | Mar 1, 2023 | Anton 2, Science Highlights
Comparison of where D-serine (D-Ser, teal dots) and glutamate (Glu, gray dots) stick to the GluN2AA subunit. From Remy A Yovanno Tsung Han Chou Sarah J Brantley Hiro Furukawa Albert Y Lau (2022) Excitatory and inhibitory D-serine binding to the NMDA receptor eLife...







